Hot topic 1: Structure determination using X-ray FELs
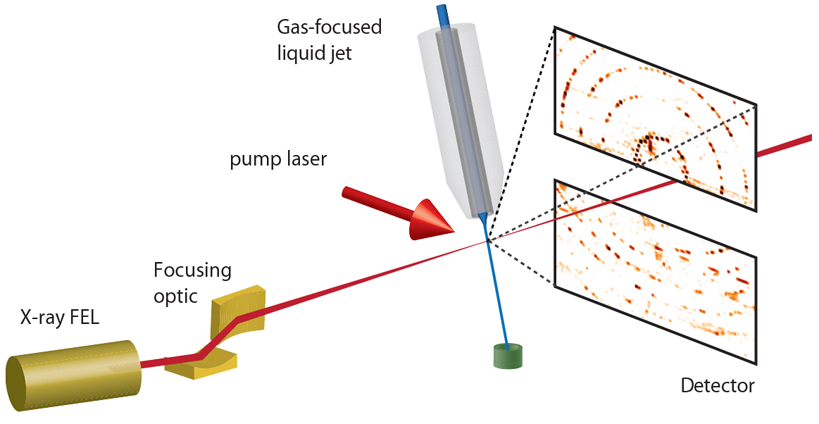
X-ray free-electron lasers offer a route to obtain structures of macromolecules at non-cryogenic temperatures without limitations of radiation damage, from small crystals. There are now five hard X-ray free-electron lasers operating in the world, and more to come, all providing instrumentation to carry out crystallography. The short, intense X-ray pulses pass through these samples before they have time to respond to the interaction, carrying information about their pristine state towards the diffraction detector. However the sample is subsequently vaporised, allowing only one flash X-ray diffractogram per crystal and necessitating an approach to build a complete dataset from many thousands of such snapshots. This method of serial crystallography especially requires the means to rapidly feed fresh crystals to the beam at the fast arrival rate of Xray pulses, a detector that can record diffraction patterns at that rate, and software to process the diffraction data to provide an accurate and complete list of structure factors. The method can very naturally be applied to obtain a time series of structures that evolve in time. In this Hot Topic session we will introduce the method, provide a live demonstration of sample injection, and offer a tutorial in the data analysis and processing using the software suite CrystFEL. We hope this will encourage many to further explore, utilise, and contribute to the method.
Part 1: An Introduction to Serial femtosecond crystallography
Henry Chapman, CFEL DESY
How do X-ray free-electron lasers provide beams that are a billion times brighter than at synchrotron radiation facilities? How can experiments be carried out with such powerful and destructive beams? What are the characteristics of flash diffraction data from small protein crystals? What are the sample requirements for a serial femtosecond crystallography experiment? What types of time-resolved measurements can be carried out? What phasing methods are available? These and many other questions will be addressed in the introductory lecture. Charting the development of the technique from the first serial serial crystallography experiment at the Linac Coherent Light Source (LCLS) in California in 2009, through many developments and improvements in experimental design and analysis, we will examine some case studies of structures that were obtained from tiny crystals. Small crystals are extremely well suited to time-resolved measurements. For photoactive proteins, reactions can be triggered by a pulse from a visible-light laser which is precisely timed to arrive a prescribed time before the measurement. The small crystals are well matched in size to the penetration depth of the optical light, ensuring that the entire volume responds to the light rather than just a small fraction. Another way to measure the response of proteins is to mix small crystals with a reactant or binding partner just prior to the exposure by the X-ray pulse. The mixed solution can be directed as a free liquid jet across the X-ray beam. Again, small crystals offer an advantage since the time for molecules to diffuse through the entire crystal can be less than 1 ms for a 1-μm-wide crystal compared with many minutes for much larger crystals. Reaction kinetics can be tracked in serial crystallography by collecting diffraction data at different distances in the flow after the mixing point. Small crystals also provide interesting effects in the diffraction measurement that may give new ways to phase diffraction data.
References
- Y. Kang et al. “Crystal structure of rhodopsin bound to arrestin by femtosecond x-ray laser,” Nature, 523 561 (2015). doi: 10.1038/nature14656
- K. Pande et al. “Femtosecond structural dynamics drives the trans/cis isomerization in photoactive yellow protein,” Science, 352, 725 (2016). doi: 10.1126/science.aad5081
- J. R. Stagno et al. “Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography,” Nature, 541, 242 (2017). doi: 10.1038/nature20599
Part 2: Hands-on Sample delivery
Saša Bajt, Photon Science DESY
The intense X-ray pulses from FELs completely destroy the sample, but not before a flash diffraction pattern can be acquired. This approach of “diffraction before destruction” requires the a fresh crystal to be delivered to the beam in time for the next pulse and measurement. Since samples do not need to be cooled to reduced radiation damage, measurements can be made at physiological temperatures (or even over a range of temperatures). This has allowed the delivery of macromolecular crystals in their crystallisation medium, such as the fluid mother liquor or the lipid cubic phase. At sources such as the LCLS, where 120 pulses are generated per second, fresh sample must pass the X-ray interaction point every 8.3 ms. However, at sources such as the European XFEL which generates bursts in which pulses arrive at rates up to 4.5 MHz, samples must pass by much faster. For example, sample must move by about 50 μm between pulses spaced by 220 ns, meaning a speed over 220 m/s. These different requirements have spurred the exploration and development of a large variety of methods to deliver crystals to XFEL beams, including free-flowing liquid jets, extrusion of lipidic cubic phase, the scanning of crystal-coated membranes and moving tapes. Other considerations for the delivery method are the sample consumption or efficiency, amount of background scattering it generates, and the size of the crystals it can carry.
In this hands-on session we will demonstrate the formation of a microjet of a crystal solution using a double flow-focussed jet. Such a liquid jet is fast enough for megahertz sources and yet has a moderate consumption of several microliters per minute of flow (at such sources, structures can be obtained in 10 minutes or so). This is achieved by forming a jet with a fluid chosen for its properties of surface tension and viscosity, into which the sample is introduced. The jet will be observed under a microscope, to see how it responds to different flow parameters and sample conditions.
References
- M. Wiedorn et al. “Megahertz serial crystallography,” Nature Comm. 9, 4025 (2018). doi: 10.1038/s41467-018-06156-7
- U. Weierstall et al., “Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography,” Nature Comm. 5, 3309 (2014). doi: 10.1038/ncomms4309
- D. Oberthuer et al., “Double-flow focused liquid injector for efficient serial femtosecond crystallography,” Sci. Rep. 7, 44628 (2017). doi: 10.1038/srep44628
Part 3: CrystFEL Tutorial
Thomas White, CFEL DESY
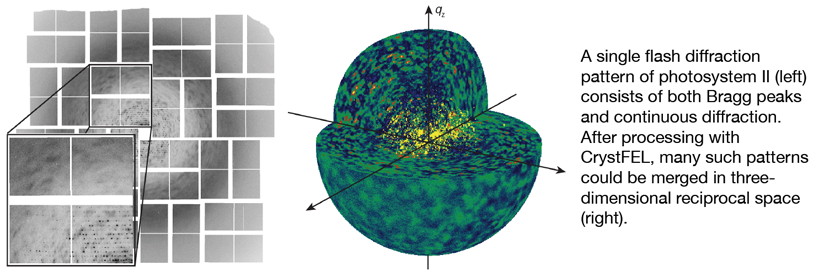
The final piece of the puzzle is to process the data and then solve the structure or calculate electron density maps to answer the biological question at hand. Because the crystals have random orientations within the liquid jet, processing starts with determining the orientation of each crystal based on the spots in the diffraction pattern. This information can then be used to calculate the locations of all spots on the detector, including those which are too weak to be immediately seen. The intensities of all the spots can then be measured, and the measurements from each of the myriad crystal diffraction patterns combined together to produce the final data file which can then be used like data from a conventional crystallographic experiment. Along the way, several properties intrinsic to the “serial” mode of data collection make matters more complicated, but also more interesting.
In this session, we will look at how to process the data from an XFEL experiment. We will cover important aspects of crystallographic theory which underpin the data processing, along with hands-on processing of some real XFEL diffraction data using the free and open-source CrystFEL software developed in our group.
References
- T. A. White. “Processing serial crystallography data with CrystFEL: a step-by-step guide”. Acta Cryst. D75 (2019). doi:10.1107/S205979831801238X
- T. A. White, R. A. Kirian, A. V. Martin, A. Aquila, K. Nass, A. Barty and H. N. Chapman. “CrystFEL: a software suite for snapshot serial crystallography”. J. Appl. Cryst. 45 (2012), p335–341. doi:10.1107/S0021889812002312
- W. Liu, D. Wacker, C. Gati, G. W. Han et al. “Serial Femtosecond Crystallography of G Protein-Coupled Receptors”. Science 342 (2013) p1522. doi:10.1126/science.1244142