Hot topic 3: Protein structure determination by NMR
Oliver Zerbe, University of Zurich, Department of Chemistry, Switzerland
Part 1: Introduction
Nuclear magnetic resonance spectroscopy (NMR) presents the second, often less well-known method for determining structures of proteins in solution. The technique was pioneered by Kurt Wüthrich, and his contribution to the field was awarded with the Nobel Prize in 2002. NMR in principle can determine structure of proteins in solution composed of arbitrary compositions of buffers, and hence can be adapted to physiological conditions. Specific isotope labeling methods in combination with 3D NMR techniques helped to push the size limits for proteins under study to the 50 kDa range, but structures at that end of the size range require special biochemical and spectroscopic efforts. NMR is not only capable of determining structures but can also provide insight into protein dynamics, but delivering order parameters, and rates for transitions between different conformations.
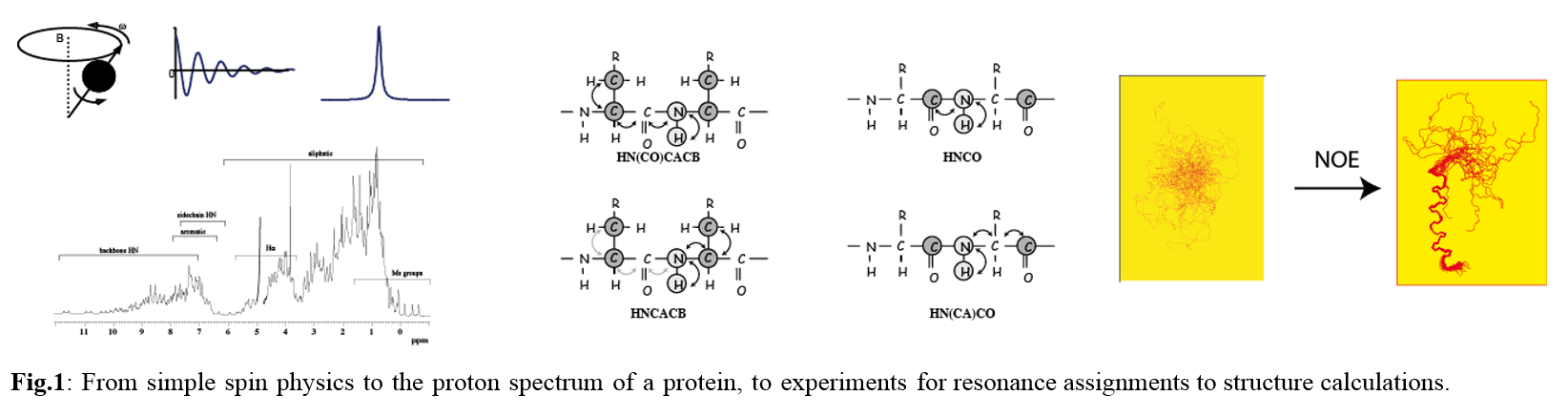
A self-consistent introduction into protein structure determination using NMR-derived data will be provided; the principle of the NMR experiments will be briefly explained and structure determination of small soluble proteins presented.
The various biochemical methods that help with the assignments of larger proteins will be discussed. The problems that are encountered when working with membrane proteins will be tackled. The various types of membrane mimics used nowadays will be considered. Finally, the methods used to study activation of GPCRs by NMR will be discussed.
References
- Chapter 6 in I. Campbell: Biophysical Techniques (Oxford Press) (an excellent book for all biophysical techniques)
- D. Marion, An Introduction to Biological NMR Spectroscopy, Mol. Cell. Proteomics, 2013, 12, 3006-3025
- K. Wüthrich, NMR Studies of Structure and Function of Biological Macromolecules (Nobel Lecture), Angew. Chem. 2003, 42,3340-3363
Part 2: Case studies: GPCR ligand complexes – agonists, antagonists and allosteric modulators
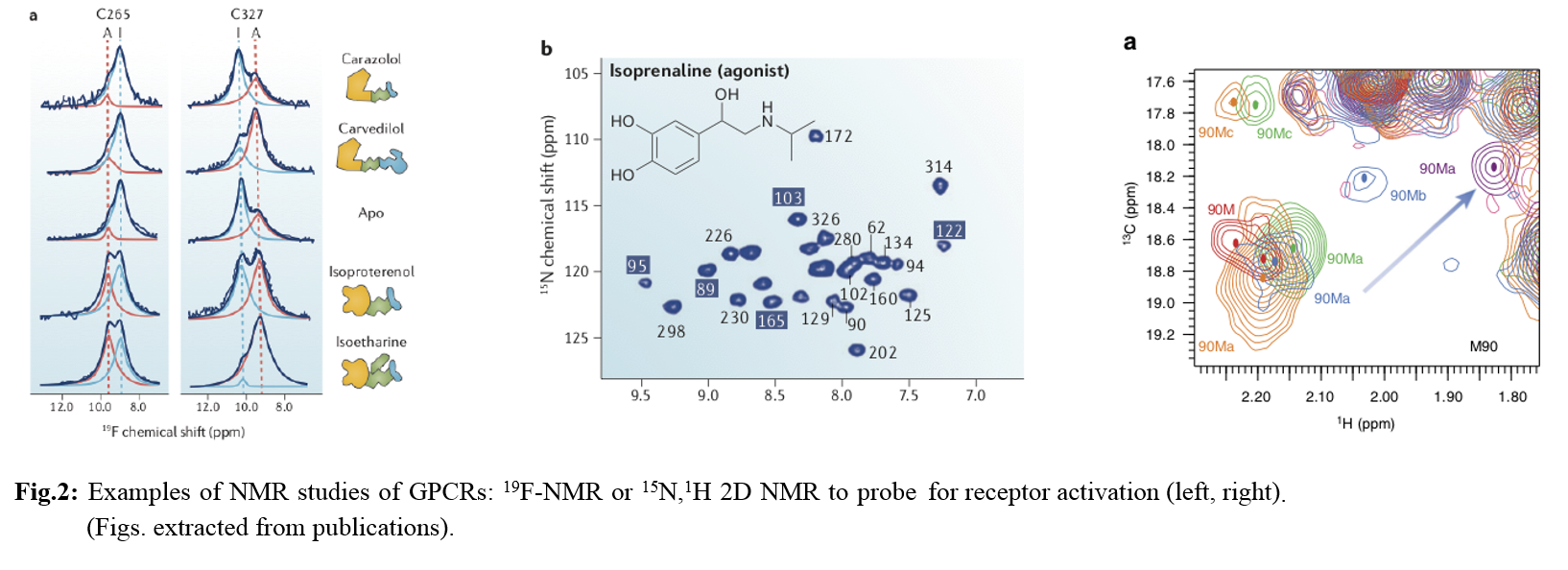
G-protein coupled receptors are pharmacologically relevant targets that due to their mode of action are inherently conformationally unstable, and therefore require substantial modifications to become amenable to crystallization trials. Since NMR does not require formation of single crystals, and because in principle it can handle partly flexible proteins it should be an ideal method to investigate GPCRs. However, the classical methodology of solving structures failed to be applicable to GPCRs so far, and it is the aim of this workshop to explain alternative methods and how NMR methods are adapted to the specific issues with membrane proteins and GPCRs in particular.
Case studies are drawn from literature comprising β1, β2 and A2A adrenergic receptors. In addition, I will show data on the α1β adrenergic receptor that is studied in my lab, as well as bacterial rhodopsins as model systems.
After the general introduction of the methods one should in the end understand why that particular type of methodology was used to address studies of activation or allosteric modulation. The participants should also comprehend what kind of information can be derived from NMR studies of GPCRs and how much work that is. NMR and X ray techniques may very well complement each other in such studies, but for a successful collaboration between crystallographers and NMR spectroscopists from my point a more detailed understanding of the technique and the particular strengths but also problems in that field are necessary.
References
- Shimada et al., GPCR drug discovery: Integrating solution NMR data with crystal and cry-EM structures, Nat. Rev. Drug. Discov. 2019, 18, 59-82
- Eddy et al., Allosteric Coupling of Drug Binding and Intracellular Signaling in the A2A Adenosine Receptor, Cell, 2018, 172, 68-80
- Osawa et al., Functional dynamics of proteins revealed by solution NMR, Core. Op. Struct. Biol., 2012, 22, 660-669
- Solt et al., Insight into partial agonism by observing multiple equilibria for ligand-bound and Gs-mimetic nanobody-bound β1-adrenergic receptor, Nat. Comm., 8, 1795
- Marino et al, Understanding GPCR Recognition and Folding from NMR Studies of Fragments, RSC Advances, 2018, 8, 9858 – 9870.
Part 3: Hands on- interactively formulate a hypothetical research proposal on GPCRs studied by NMR
Finally, I want to put together a research proposal for a project that aims at performing NMR studies of a GPCR following suggestions from the audience. We will discuss what to do in that project, what type of information we want to derive, and pay particular attention to the experimental part, the problems that might (will!) show up during the project. A thorough discussion with the attendants will help them to understand the technique in much more detail, and in particular recognize the scope of the project and the particular aspects that must be dealt with to become successful.