Maximilian Edich1 , David Briggs2 , Gianluca Santoni3, Andrea Thorn1
1Institut für Nanostruktur und Festkörperphysik at Hamburg University, Hamburg, Germany
2Francis Crick Institute, London, United Kingdom
3European Synchrotron Radiation Facility, Grenoble, France
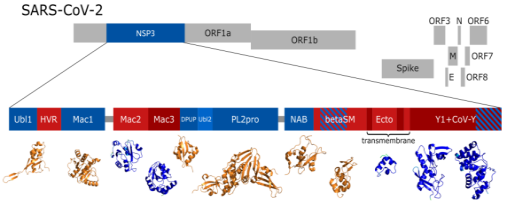
Non-structural protein 3 (nsp3) plays an essential role in the life cycle of SARS-CoV-2 and with 16 domains and roughly 2000 residues it is the largest protein of the virus. However, it is difficult to obtain an atomic resolution structure of the whole protein due to several regions of intrinsic disorder and transmembrane domains. The numerous 3D models deposited in the PDB cover only 8 out of 16 domains, leaving many questions open (see Figure 1). To address this issue, we previously evaluated these structures in order to establish their quality and interplay.1,2 Here, we contribute to a complete picture by utilizing the structure prediction software AlphaFold23 and bioinformatical analysis to investigate all of its domains together. Our approach revealed a previously undescribed domain and hinted towards the function of the nidovirus-wide conserved domain Y1. We followed this up with subsequent validation experiments. Our method is applicable to other multi-domain proteins and demonstrates how structural bioinformatics and AI-based fold prediction complement traditional experiments in structural biology.
References:
[1] Croll, Tristan I. et al. “Making the invisible enemy visible.” Nature Structural & Molecular Biology 28.5 (2021): 404-408.
[2] von Soosten, Lea C., et al. “The Swiss army knife of SARS-CoV-2: the structures and functions of NSP3.” Crystallography Reviews 28.1 (2022): 39-61.
[3] Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596 (2021), 583–589.
This work was supported by the German Federal Ministry of Education and Research [grant no. 05K19WWA and 05K22GU5] and the Deutsche Forschungsgemeinschaft [grant no. TH2135/2-1].
Keywords: SARS-CoV-2; AlphaFold2; multidomain; Nsp3; structural biology
