


Activity A1: Bring your sample
- Having troubles crystallizing your protein/nucleic acid/complex?
- Getting crystals, but they are desperately poor, don’t diffract, are difficult to optimize, or require a different morphology or space group?
- Willing to try an inovative, in-depth and scientifically sound combination of methods?
YES, YES and YES?
Then apply online to take part in this activity by simply completing the online form (only once you are registered for the HTTC5), wait for our feedback, and if it is a YES, then bring your own sample, and take part in the Activity A1: Bring your sample at the HTCC5, organized by Croatian Association of Crystallographers, Douglas Instruments & CrystalImpact!
As for the sample, we’ll need a minimum of 10 microLIT, ideally 30 microLIT of the sample solution. If you happen to already have those poor crystals, and/or crystal of a structurally related macromolecule, and are willing to bring them too, that’d be just awesome. If not, bring the solution, we’ll manage.
The activity, which will run over the entire workshop, starting with sample collection right upon registration, will include the opportunity for students to bring their own protein samples, prepared to be crystallized, but reluctant to produce a desired outcome, to learn important techniques for sample preparation and extend knowledge about their own samples by combining preparation and analytical methods.

The training will include the opportunity to carry out solubility screening and microseeding [1] experiments on an automatic system, which can increase the likelihood of obtaining diffracting crystals. The technique is also useful if you have crystals of a related protein. Cross seeding [2] is becoming routine and can be used as long as there is some homology between the protein and the seed crystals. Cross seeding is great for:
- Seeding into complexes
- Seleno methylamine or heavy atom derivatives
- Seeding into Different constructs or species (see second reference below)
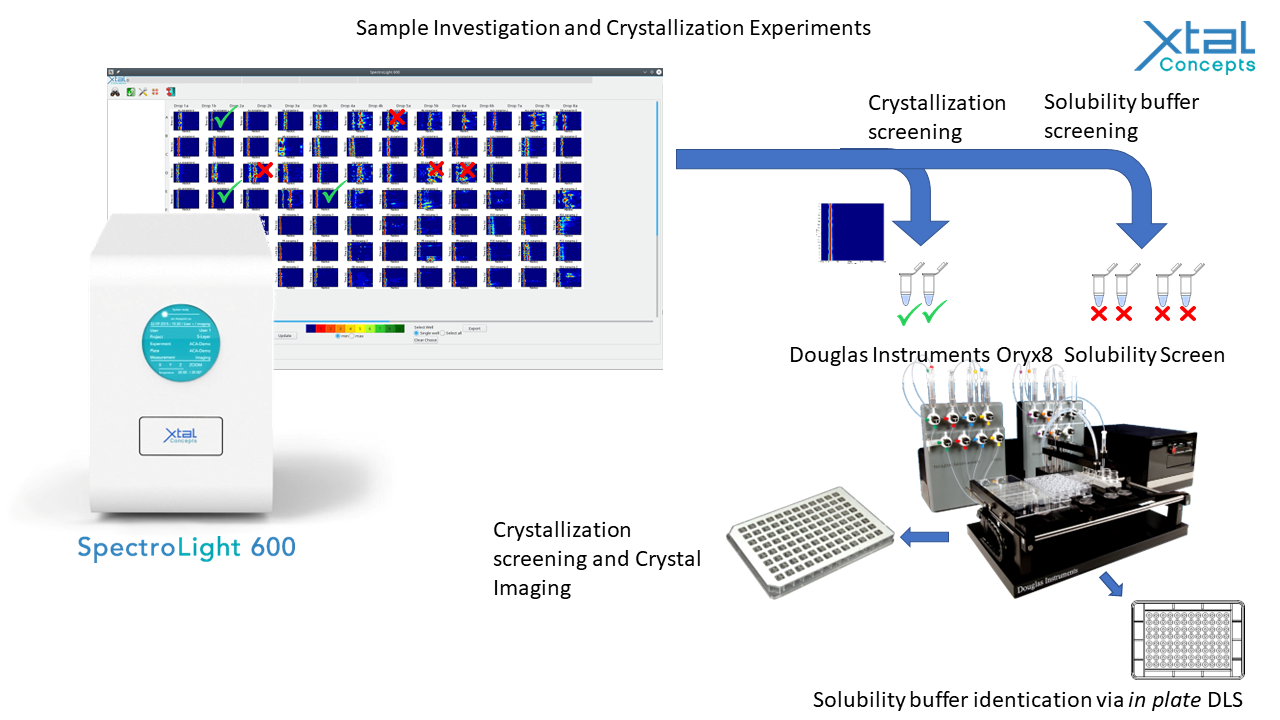
Students will be taught how to make seed-stocks and given guidance for setting up crystallization experiments. They will also be shown how to set up microbatch-under-oil experiments, which are easier to scale up to make samples for serial data-collection, cryoEM, neutron diffraction etc. Sample investigation before and after screening will focus on sub-microliter in plate dynamic light scattering (DLS) providing a deeper understanding of sample formulation and screening. All collected data will be summarized and shared with students due to the fundamental significance of such data and for enabling utilization beyond the event. The main practical session (18/04/2022, 18:30 – 19:30) will show the methods being applied to a few samples on-screen to all delegates. Other experiments can be set up and advice given during the coffee and lunch breaks.
Important: We won’t be able to fit in samples from all delegates, so number will be limited to approximately 10 samples. If you intend to bring your own sample for this activity, please do complete and submit the online sample questionnaire. Based on your answers we shall choose up to ten samples best suited for this activity.
SESSIONS:
- 16/04/2022, Registration: All students invited to bring their own samples will hand over the samples on the registration desk
- 16/04/2022, 18:30 onwards: Samples will be subjected to initial DLS analysis and crystallization experiments will be setup
- 17/04/2022: 14:30 – 15:30: Theoretical background of the Activity A1, followup on the samples put to crystallize
- 18/04/2022: 18:30 – 19:30: Onscreen Experiment demonstration, followup on the samples put to crystallize
The practical sessions will be coordinated by Douglas Instruments and XtalConcepts, who are kindly making equipment available for the workshop.
REFERENCES:
[1] D’Arcy, Allan, Frederic Villard, and May Marsh. “An automated microseed matrix-screening method for protein crystallization.” Acta Crystallographica Section D: Biological Crystallography 63.4 (2007): 550-554.
[2] Abuhammad, A., Lowe, E.D., McDonough, M.A., Shaw Stewart, P.D., Kolek, S.A., Sim, E. and Garman, E.F., 2013. Structure of arylamine N-acetyltransferase from Mycobacterium tuberculosis determined by cross-seeding with the homologous protein from M. marinum: triumph over adversity. Acta Crystallographica Section D: Biological Crystallography 69.8 (2013): 1433-1446.
