T4: CCP4 Cloud Tutorial – Refinement with REFMAC5, Ligand Dictionary Generation with AceDRG, and Interactive Model Building & Ligand Fitting with Coot
HT2: Processing
T4 (Tutorial): Interactive Model Building, Ligand Fitting and Refinement with Coot
Rob Nicholls
MRC Laboratory of Molecular Biology
Cambridge Biomedical Campus
Cambridge, UK
CCP4 Cloud (Krissinel et al., 2022) includes three in-built MX refinement tutorials:
- Simple refinement – which focusses on adjustment of data-geometry weighting and inspection of refinement results.
- Twin refinement – which demonstrates how to extract information about twinning from REFMAC5 output.
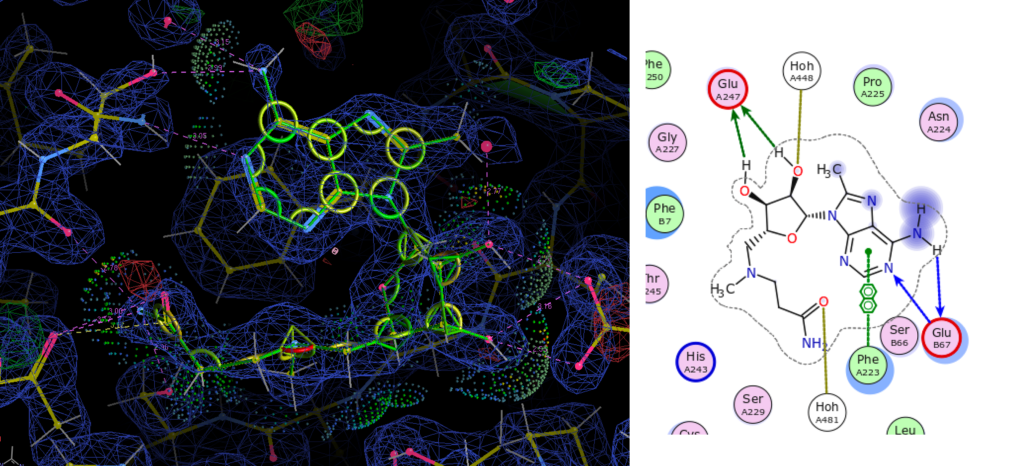
- Ligand Dictionary Generation, Fitting and Refinement – which uses AceDRG to generate a ligand dictionary and conformer, Coot to fit the ligand into the macromolecular model, REFMAC5 to refine the protein-ligand complex model, and finally Coot to validate the refined ligand binding site.
It should be noted that the CCP4 suite only includes these tutorials if they were selected during installation. Otherwise, the CCP4 Cloud tutorials will only be available when using the “remote” version of CCP4 Cloud (ccp4cloud-remote), which uses the CCP4 server for project management and processing.
In this session we shall focus on working through the third tutorial and discussing the procedure of ligand dictionary generation, fitting, refinement and validation in CCP4. More information on this topic can be found in Nicholls (2017).
- Krissinel, E. et al. (2022) CCP4 Cloud for structure determination and project management in macromolecular crystallography. Acta Cryst. D78, 1079-89.
- Nicholls (2017) Ligand fitting with CCP4. Acta Cryst. D73, 158-70.
|


